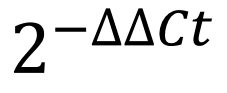
The delta-delta Ct method, also known as the 2 – ∆∆Ct method, is a simple formula used in order to calculate the relative fold gene expression of samples when performing real-time polymerase chain reaction (also known as qPCR). The method was devised by Kenneth Livak and Thomas Schmittgen in 2001 and has been cited over 61,000 times.
Further video tutorials on qPCR data analysis can be found in our Mastering qPCR course
>>Use code 20QPCR to get 20% off
We have created a FREE Excel template which contains all of the formula described in this article below. Use this to practise and get the hang of the calculations. Everything is done for you, all that is required is the Ct values!
If you would like to download this, simply click here.
It is worthwhile understanding what the delta-delta Ct formula means before diving straight into the calculations.
The overall formula to calculate the relative fold gene expression level can be presented as:
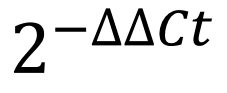
This looks like a scary mathematical formula when in actual fact, it isn’t. Let’s break the formula down into easier to understand chunks.
Firstly, Ct stands for the cycle threshold (Ct) of your sample. This is given after the qPCR reaction by the qPCR machine. Simply, it is the cycle number where the fluorescence generated by the PCR produce is distinguishable from the background noise.
The symbol ∆ refers to delta. Delta is a mathematical term used to describe the difference between two numbers. So it is useful to use when summarising long formulas.
So, let’s take a look to see what the ∆∆Ct part of the equation means:
∆∆Ct = ∆Ct (treated sample) – ∆Ct (untreated sample)
Essentially, ∆∆Ct is the difference between the ∆Ct values of the treated/experimental sample and the untreated/control sample. But what does ∆Ct refer to?
Let’s take a look:
∆Ct = Ct (gene of interest) – Ct (housekeeping gene)
Basically, ∆Ct is the difference in Ct values for your gene of interest and your housekeeping gene for a given sample. This is to essentially normalise the gene of interest to a gene which is not affected by your experiment, hence the housekeeping gene-term.
To use the delta-delta Ct method, you require Ct values for your gene of interest and your housekeeping gene for both the treated and untreated samples. If you have more than one housekeeping gene, it may be worth checking out the guide on analysing qPCR data with numerous reference genes.
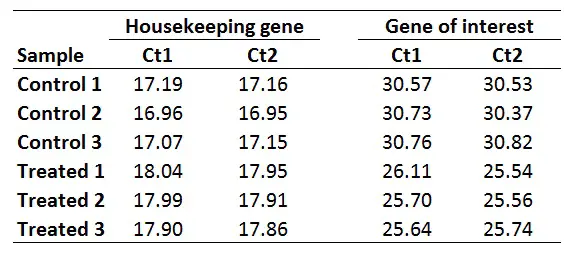
Here is how to calculate the relative gene expression in 5 easy steps.
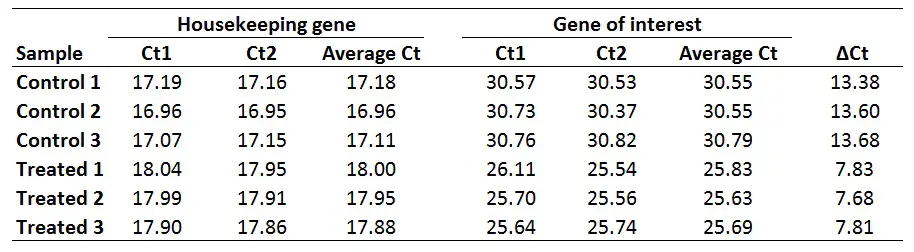
The first step is to average the Ct values for the technical replicates of each sample. So, when performing the qPCR in duplicate or triplicate, for example, these values need to be averaged first. In the example below, each sample was run in duplicate (Ct1 and Ct2).
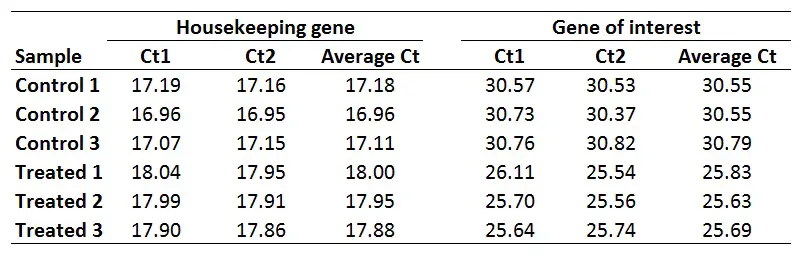 2. Calculate the delta Ct for each sample
2. Calculate the delta Ct for each sampleThe next step is to calculate delta Ct (∆Ct) for each sample by using the newly created average Ct values. The formula to calculate delta Ct is presented below.
∆Ct = Ct (gene of interest) – Ct (housekeeping gene)
For example, to calculate the ∆Ct for the ‘Control 1‘ sample:
∆Ct Control 1 = 30.55 – 17.18
The next step is to decide which sample, or group of samples, to use as a calibrator/reference when calculating the delta-delta Ct (∆∆Ct) values for all the samples. This is the part which confuses a lot of people. Basically, this all depends on your experiment set-up.
A common way of doing this is to just match the experimental samples and determine the relative gene expression ratios separately. This is all well and true for experiments that have matched pairs, such as the case in cell culture experiments. However, this is difficult when the two experimental groups vary in n numbers and do not have matched pairs.
Another way to select a calibrator/reference sample is to pick the sample with the highest Ct value, so the sample with the lowest gene expression. This way, all the results will be relative to this sample. Or, you could simply select just one of the control samples to act as the reference sample.
I personally average the ‘Average Ct’ values of the biological replicates of the control group to create a ‘Control average’. By doing so would mean that the results are presented relative to the control average Ct values.
Whichever sample, or group of samples, you use as your calibrator/reference is fine so long as this is consistent throughout the analyses and is reported in the results so it is clear. Remember, the results produced at the end are relative gene expression values.
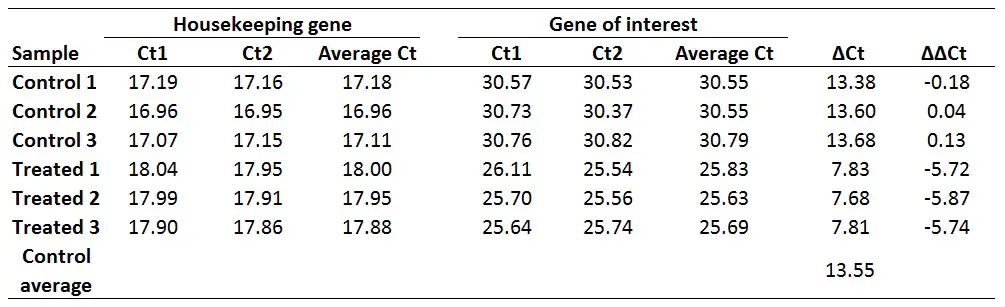
With this in mind, if we want to get ∆∆Ct values for every sample (including for each control sample), we first need to average the ∆Ct for the 3 control samples:
∆Ct Control average = (13.38 + 13.60 + 13.68)/3

Note, if the Ct values are variable, then it may be more appropriate to use the geometric mean instead of the arithmetic mean above. The geometric mean is more resistant to outliers, compared with the arithmetic mean. The geometric mean is used in the Vandesompele relative gene expression method for this reason.
For example, if the Ct values for my three control samples were 13.38, 13.60 and 15.80 instead, then this is a good reason to use the geometric mean rather than the arithmetic mean.
To use the geometric mean, firstly multiply the numbers together and then take the nth root of that value. The n is simply the number of observations in the formula, which is 3 in this example. So, using my latest example, this would be:
∆Ct Control geometric average = ∛(13.38 x 13.60 x 15.80)
Now calculate the ∆∆Ct values for each sample. Remember, delta delta Ct values are relative to the untreated/control group in this example. The formula to calculate delta delta Ct is presented below.
∆∆Ct = ∆Ct (Sample) – ∆Ct (Control average)
For example, to calculate the ∆∆Ct for the Treated 1 sample:
∆∆Ct Treated 1 = 7.83 – 13.55
Finally, to work out the fold gene expression we need to do 2 to the power of negative ∆∆Ct (i.e. the values which have just been created). The formula for this can be found below.
Fold gene expression = 2^-(∆∆Ct)
For example, to calculate the fold gene expression for the Treated 1 sample:
Fold gene expression = 2^-(-5.72)
Doing this would give a fold gene expression of 52.71 for the Treated 1 sample. Doing this for all of the samples will look like this:
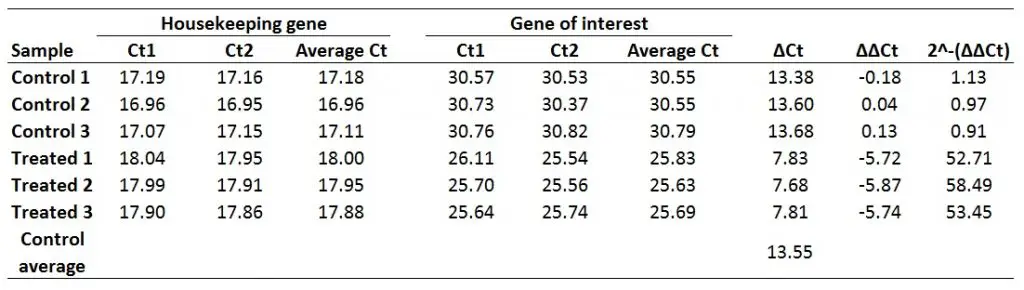
And that is how you can use the delta-delta Ct method to work out the fold gene expression for your samples.
Just a point regarding statistical analysis of the gene expression values. It is always best to log transform the values (2^-∆∆Ct) before undertaking statistical analysis. This is because the untransformed gene expression values will most likely not be normally distributed and heavily skewed, especially in experiments where a strong stimulus is used. To do log transformations in Excel, simply use the log formula (=Log).
Then, the choice of statistical test will be dependent on your experimental set-up. If you are struggling to perform a particular test, refer to our selection of SPSS and GraphPad Prism tutorials.
Steven is the founder of Top Tip Bio. He is currently a Medical Writer and a former Postdoctoral Research Associate. Enjoyed the tutorial? Then let me know by leaving a comment below, or consider buying me a coffee.